Medical Image Analysis
What is Medical Image Analysis?
Medical images are the images that are taken when you have a medical checkup or examination at a hospital. These include X-ray images, Computed Tomography (CT) images, Magnetic Resonance Image (MRI) images, ultrasound (echo) images, endoscopic images (gastroscopy, etc.), pathological images, dermatological images, and fundus images. Medical image analysis is the process of extracting information from these medical images using machine learning and other technologies to identify the location of lesions and classify lesions. Medical image analysis has been actively studied for a long time, and after the development of deep learning, medical image analysis technology has been rapidly advancing along with general image analysis technology.
Each medical image has its own characteristics, and these characteristics often pose challenges for machine learning analysis. For example, most images are two-dimensional, but CT and MRI images are three-dimensional images consisting of two-dimensional tomographic images stacked on top of each other. Endoscopic images are two-dimensional, but since they show the inside of the digestive tract, they have a three-dimensional extent. A pathological image is a microscopic image of cellular tissue, etc., consisting of tens of thousands of pixels by tens of thousands of pixels, and has a very large resolution.
Thus, medical image analysis is not simply a matter of training a machine learning model; it must be tailored to the characteristics of the image.
Medical images
Related works
The most famous technology to emerge from medical image analysis is a semantic segmentation network called U-Net [1]. This network is named after its U-shaped arrangement of encoders and decoders. Its simple structure and lightweight parameters have made it one of the most popular semantic segmentation models in the field of general imaging. It is still used in medical image analysis, where it is difficult to collect a large number of images.
Since the advent of deep learning, there have also been many reports of performance comparable to that of physicians. For example, H. A. Haenssle et al. used the International Skin Imaging Collaboration (ISIC) database to build a model that can determine whether a skin image is a mole or malignant melanoma, achieving accuracy comparable to that of a dermatologist [2]. V. Gulshan et al. constructed a model to determine whether a patient has diabetes using 128,175 fundus images (images of the retina and other parts of the eye), and achieved accuracy comparable to that of ophthalmologists [3]. In pathological images, a competition was held for the task of detecting lymph node metastasis in breast cancer, and it was reported that accuracy exceeding that of physicians was achieved in some tasks where there was a time limit [4, 5].
Machine learning techniques have been used to approach various lesions in this way, and recently, many studies have been conducted to diagnose COVID-19 (coronavirus) infection from CT images [6, 7, 8].
Uniqueness and Achievements
In 2017, our laboratory participated in a project on the Medical Image Big Data Cloud Infrastructure[9] of the National Institute of Informatics Medical Big Data Research Center with support from the Japan Agency for Medical Research and Development (AMED). Since many medical societies participated in this project, a large number of images were collected on the cloud infrastructure. Although it has been difficult to collect a large number of medical images, this project has made it possible to build a machine learning model using a large number of images [9]. Prof. Harada received the Commendation for Science and Technology by the Minister of Education, Culture, Sports, Science, and Technology, Prize for Science and Technology (Promotion Category) for his contribution to the research on AI automatic diagnosis on the cloud platform.
Below is an example of our research conducted using the medical big data cloud platform.
Pathological Image Analysis
In pathology images, we need to deal with images that have a side of 10 to the fifth power of a pixel and a size of several gigabytes per image. In other words, we have to deal with images that are typically displayed on a personal computer, scaled up by a factor of 100 in height and width. In conventional research, large images are handled by dividing them into patches (portions), analyzing each of them, and combining the results. However, this method cannot adequately take into account the relationships between batches. It does not handle global information. We proposed a new model for the task of detecting anomalies in pathology images that takes into account global information and achieved state-of-the-art performance in 2019. The results have been presented at a top-level international conference called ICCV [10]. Based on this method, we also performed further analysis on gastric biopsy images in collaboration with pathologists and showed that our method has high accuracy even on data from other institutions, which is generally considered to be less accurate for pathology images [11].
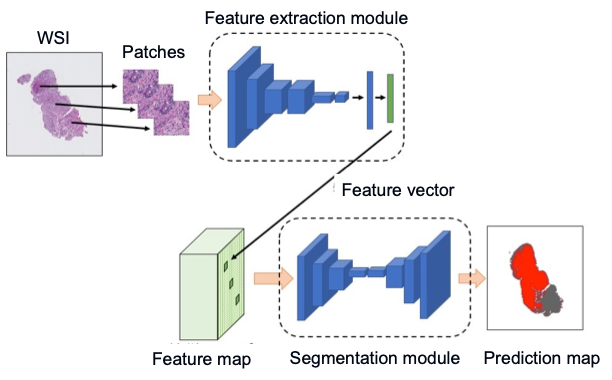
Overview of the proposed method
Domain adaptation methods are also being developed to cope with differences in different organs. In pathology, it is said that images from different organs are completely different. For example, the accuracy of inference on a colon biopsy image is greatly reduced if the model is trained on a stomach biopsy image. Therefore, we proposed a learning method that combines multiple instance learning and domain adaptation to deal with ultra-high-resolution pathology images, and demonstrated the superiority of the proposed method through experiments. The results have been presented at ISBI, a conference on medical imaging systems [12].
Future Directions
Our laboratory currently has staff and students specializing in medical imaging. We have many opportunities to listen to the findings of doctors and experts and are engaged in advanced and practical research using our own datasets. In the future, we expect to deepen our discussions with doctors and also advance research closer to application. In fact, the model we proposed at ICCV in 2019 was endorsed by physicians as having practical utility, and we have filed a patent application.
References
[1] O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 2015, vol. 9351, pp. 234–241. [2] H. A. Haenssle et al., “Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists,” Ann. Oncol., vol. 29, no. 8, pp. 1836–1842, Aug. 2018. [3] V. Gulshan et al., “Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs,” JAMA, vol. 316, no. 22, pp. 2402–2410, Dec. 2016. [4] B. Ehteshami Bejnordi et al., “Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer,” JAMA, vol. 318, no. 22, pp. 2199–2210, Dec. 2017. [5] P. Bándi et al., “From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge,” IEEE Trans. Med. Imaging, vol. 38, no. 2, pp. 550–560, 2019. [6] H. X. Bai et al., “AI Augmentation of Radiologist Performance in Distinguishing COVID-19 from Pneumonia of Other Etiology on Chest CT,” Radiology, vol. 78, no. May, p. 201491, Apr. 2020. [7] T. Uemura, J. J. Näppi, C. Watari, T. Hironaka, T. Kamiya, and H. Yoshida, “Weakly unsupervised conditional generative adversarial network for image-based prognostic prediction for COVID-19 patients based on chest CT,” Med. Image Anal., vol. 73, p. 102159, 2021. [8] I. Dayan et al., “Federated learning for predicting clinical outcomes in patients with COVID-19,” Nat. Med., vol. 27, no. 10, pp. 1735–1743, Oct. 2021. [9] K. Murao, Y. Ninomiya, C. Han, and K. Aida, “Cloud platform for deep learning-based CAD via collaboration between Japanese medical societies and institutes of informatics,” in Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications, Mar. 2020, vol. 11318, pp. 223–228. [10] S. Takahama, Y. Kurose et al., “Multi-Stage Pathological Image Classification using Semantic Segmentation,” in The IEEE International Conference on Computer Vision (ICCV), 2019, pp. 10702–10711. [11] H. Abe, Y. Kurose et al., “Development and multi-institutional validation of an artificial intelligence-based diagnostic system for gastric biopsy,” Cancer Sci., Aug. 2022, doi: 10.1111/cas.15514. [12] S. Takahama, Y. Kurose et al., "Domain Adaptive Multiple Instance Learning for Instance-Level Prediction of Pathological Images, " in 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), 2023.